The SMARTer Human TCR a/b Profiling Kit enables users to analyze T-cell receptor (TCR) diversity from input RNA samples. This kit is designed to work with a range of RNA input amounts depending on the sample type, and has been shown to generate high-quality libraries from as little as 10 ng to 3 μg of total RNA obtained from peripheral blood leukocytes, or from 50 to 10,000 purified T cells. As the name suggests, the kit can be used to generate data for both alpha- and beta-chain diversity, either in the same experiment or separately.
This kit leverages SMART technology and employs a 5' RACE-like approach to capture complete V(D)J variable regions of TCR transcripts. Included in the kit are primers that incorporate Illumina-specific adaptor sequences during cDNA amplification. The protocol generates indexed libraries that are ready for sequencing on Illumina platforms.
- Complete V(D)J sequence information—Obtain full-length sequences for variable regions of TCR mRNA transcripts.
- TCR-alpha and TCR-beta—Profile diversity for both TCR subunits, either in the same experiment, or separately.
- Flexible sample inputs—Start from 10 ng–3 µg of RNA obtained from peripheral blood, or from 50–10,000 purified T cells.
- No multiplex PCR required—Amplify sequences for each TCR subunit with a single primer pair per reaction.
- Illumina-ready sequencing libraries—Incorporate Illumina adapter and index sequences in a ligation-independent manner, and multiplex up to 96 libraries in a single flow-cell lane.
- Exceptional sensitivity—Detect low-abundance TCR sequences, even with shallow read depths.
- Excellent reproducibility—Generate consistent results across a range of sample input amounts.
Applications
Human TCR repertoire analysis (TCR-alpha and TCR-beta subunits)
Sequencing reads on target and correlation of clonotype count data for varying sample input amounts

Sequencing reads on target and correlation of clonotype count data for varying sample input amounts. Panel A. Percentages of sequencing reads that map to CDR3 regions in either TCR-α (blue) or TCR-β (purple) or that represent off-target reads (gray). The experimental protocol was performed on three different amounts of peripheral blood RNA: 10 ng, 100 ng, and 1,000 ng. Panel B. Correlation of clonotype count data for 100 ng input RNA vs. 1,000 ng input RNA. Pearson (R) and Spearman (ρ) correlation coefficients are included.
Assessing the sensitivity and reproducibility of the SMARTer approach

Assessing the sensitivity and reproducibility of the SMARTer approach. The experimental protocol was performed in replicate on PBMC RNA samples spiked at varying concentrations (10%, 1%, 0.1%, 0.01%, and 0.001%) with RNA obtained from a homogenous population of Leukemic Jurkat T cells (TRAV8-4-TRAJ3, TRBV12-3-TRBJ1-2 clonotype). Panel A. Correlation between concentration of spiked-in Jurkat RNA and number of TRBV12-3-TRBJ1-2-specific sequence reads. Numbers along the X-axis indicate serial-diluted concentrations of spiked-in Jurkat RNA (by mass): 1 = 10%; 2 = 1%; 3 = 0.1%; 4 = 0.01%; 5 = 0.0.01. Count data for TRBV12-3-TRBJ1-2-specific sequence reads were normalized by subtracting the number of corresponding reads obtained for negative control samples consisting of unspiked PBMC RNA. Normalized count data were then Log<sub>10</sub> transformed. Circles and triangles correspond to experimental replicates for each sample concentration. Results of linear regression analysis are indicated in the upper right region of the graph. Panel B. Count data, signal-to-noise ratios, and statistical analysis for TRBV12-3-TRBJ1-2-specific sequence reads obtained from spiked RNA samples. Signal-to-noise ratios were generated using the mean counts of TRBV12-3-TRBJ1-2-specific sequence reads for each pair of experimental replicates. Rows highlighted in yellow include concentrations of spiked-in Jurkat RNA for which statistically significant elevations in TRBV12-3-TRBJ1-2-specific sequence reads were detected relative to background counts observed for unspiked negative control RNA samples.
Library preparation workflow and PCR strategy for TCR profiling using the SMARTer approach
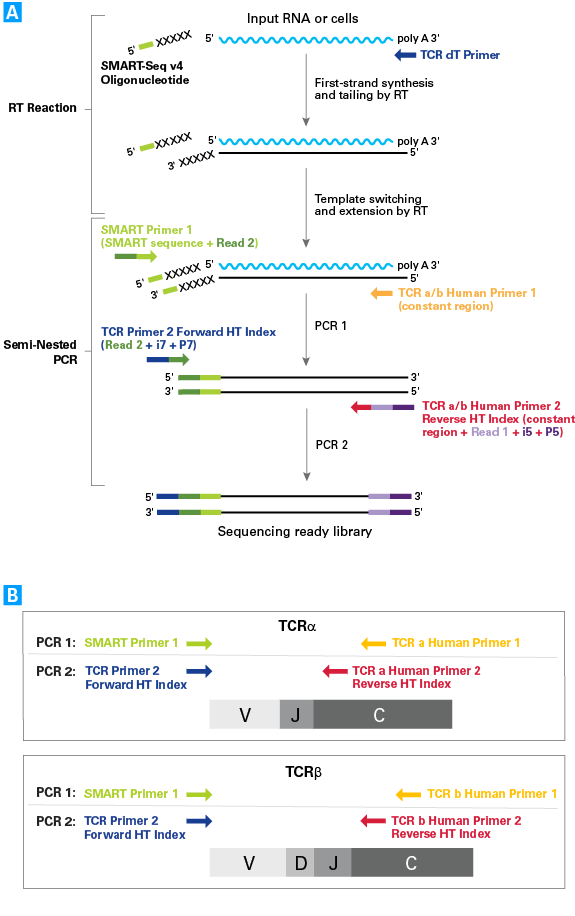
Library preparation workflow and PCR strategy for TCR profiling using the SMARTer approach. Panel A. Reverse transcription and PCR amplification of TCR subunit mRNA sequences. First-strand cDNA synthesis is primed by the TCR dT Primer and performed by an MMLV-derived reverse transcriptase (RT). Upon reaching the 5′ end of each mRNA molecule, the RT adds non-templated nucleotides to the first-strand cDNA. The SMART-Seq v4 Oligonucleotide contains a sequence that is complementary to the non-templated nucleotides added by the RT, and hybridizes to the first-strand cDNA. In the template-switching step, the RT uses the remainder of the SMART-Seq v4 Oligonucleotide as a template for the incorporation of an additional sequence on the end of the first-strand cDNA. Full-length variable regions of TCR cDNA are selectively amplified by PCR using primers that are complementary to the oligonucleotide-templated sequence (SMART Primer 1) and the constant region(s) of TCR-α and/or TCR-β subunits (TCR a/b Human Primer 1). A subsequent round of PCR is performed to further amplify variable regions of TCR-α and/or TCR-β subunits and incorporate adapter sequences, using TCR Primer 2 and TCR a/b Human Primer 2. Included in the primers are adapter and index sequences (read 2 + i7 + P7 and read 1 + i5 + P5, respectively) that are compatible with the Illumina sequencing platform. Following purification, size selection, and quality analysis, the TCR cDNA library is ready for sequencing. Panel B. Semi-nested PCR approach for amplification of TCR-α and/or TCR-β subunits. The primer pairs used for the first round of amplification capture the entire variable region(s) and most of the constant region(s) of TCR-α and/or TCR-β cDNA. The second round of amplification retains the entire variable region(s) of TCR-α and/or TCR-β cDNA, and a smaller portion of the constant region(s). The anticipated size of final TCR library cDNA (inserts + adapters) is ~700–800bp.
 Menu
Menu