- Simple workflow to generate stranded Illumina sequencing-ready libraries in 7 hours
- Appropriate for analysis of 1–1,000 intact mammalian cells or 10 pg–10 ng of low- or high-quality mammalian total RNA
- Reproducible, accurate detection of full-length coding and noncoding transcripts from single cells or total RNA
- Comparable sensitivity to our gold-standard SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing.
Interested in more data and FAQs about this product? Visit the NGS Learning Center.
Applications
- Generation of Illumina sequencing-ready libraries in 7 hours from small quantities of mammalian cells (1–1,000 cells) or total RNA (10 pg–10 ng)
- Whole transcriptome sequencing with strand-of-origin information
SMART-Seq Stranded Kit workflow
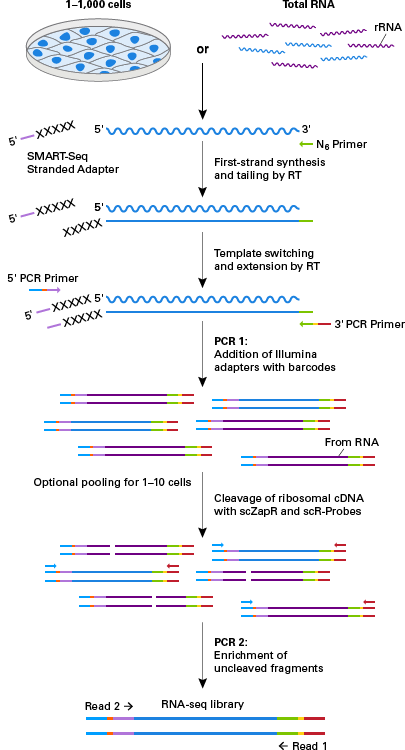
Figure 1. Schematic of the SMART-Seq Stranded Kit workflow.
SMART-Seq Stranded Kit performs well across a wide range of inputs
| Sequencing alignment metrics for A375 total RNA and cells |
| Input |
Total RNA |
1,000 cells |
500 cells |
100 cells |
10 cells |
5 cells |
1 cell |
| Number of reads (pairs) |
6,000,000 |
6,000,000 |
6,000,000 |
6,000,000 |
6,000,000 |
6,000,000 |
5,873,974 |
| Number of transcripts >1 FPKM |
13,260 |
13,294 |
13,583 |
13,520 |
12,726 |
12,602 |
11,540 |
| Number of transcripts >0.1 FPKM |
21,334 |
21,113 |
21,365 |
21,145 |
20,550 |
18,888 |
15,815 |
| Proportion of reads (%): |
| Exonic |
34.7 |
36.4 |
39.2 |
42.7 |
36.7 |
36.2 |
37.3 |
| Intronic |
29.6 |
29.3 |
27.7 |
28.3 |
34.0 |
30.4 |
21.1 |
| Intergenic |
14.2 |
13.4 |
12.2 |
12.9 |
16.7 |
16.8 |
10.1 |
| rRNA |
7.0 |
11.4 |
11.5 |
6.3 |
3.6 |
4.9 |
7.1 |
| Mitochondrial |
4.1 |
3.5 |
3.7 |
4.9 |
3.8 |
4.4 |
4.6 |
| Overall mapping (%) |
89.6 |
93.9 |
94.3 |
95.1 |
94.9 |
92.7 |
80.2 |
| Duplicate rate (%) |
37.3 |
45.2 |
40.3 |
46.1 |
52.5 |
72.2 |
78.5 |
| lncRNA mapping: |
| Number of mapped reads (%) |
7.2 |
10.4 |
10.8 |
9.4 |
8.7 |
8.6 |
7.3 |
| lncRNA transcripts detected |
5,395 |
4,687 |
4,565 |
5,439 |
5,440 |
4,983 |
2,802 |
Figure 2: High reproducibility across cell input amounts. A375 cells isolated by FACS were used to generate RNA-seq libraries with the SMART-Seq Stranded Kit. Input varied from 1 cell to 1,000 cells, with two replicates per input of 5–1,000 cells and 12 replicates for the single cells. For comparison, two aliquots of 1,000 cells were used for total RNA purification and then used for library preparation.
SMART-Seq Stranded Kit matches the sensitivity of SMART-Seq v4 technology
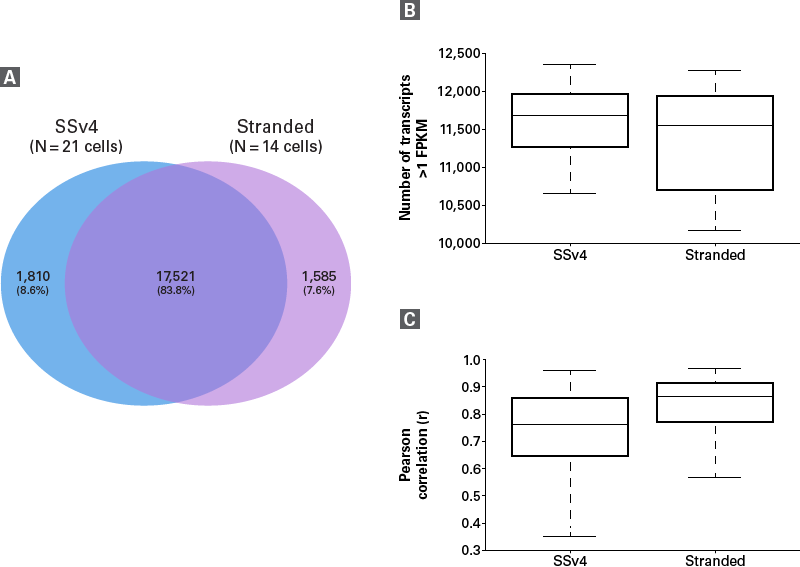
Figure 3. Comparison between SMART-Seq v4 and SMART-Seq Stranded kits. Single cells (K562) isolated by FACS were used to generate RNA-seq libraries with the SMART-Seq Stranded Kit (Stranded) and a SMART-Seq v4 Kit (SSv4; cDNA from this kit was further processed with a Nextera® XT DNA Library Preparation Kit). Panel A. The overlap in the total number of transcripts identified (FPKM >1) by each kit was analyzed and shown to be 83.8%. Panel B. The number of transcripts identified (FPKM >1) in individual cells was similar between the two kits, with a tighter range across cells processed with the SSv4 kit. Panel C. The reproducibility (Pearson correlation) of transcript expression levels across all cells from each kit was similar, although slightly higher and more consistent across cells processed with the Stranded kit.
 Menu
Menu