An important update is available for FreeStyle LibreLinkØ. Check here for more information.
 Menu
Menu
12 Mar 2020

|
Climate change impacts our health as well as our landscape. Rising temperatures can threaten the quality of the air we breathe, as well as our food and water supplies. The increased incidence of extreme weather events such as hurricanes can raise the threat of water-borne illnesses by contaminating drinking water (through exposure to soil, sewage, and infected animals), promoting the growth of pathogens and the vectors that transmit them, and increasing the likelihood of skin exposure to contaminated water sources.
Leptospirosis is a disease caused by the Leptospira bacteria that can provoke kidney damage, meningitis, liver failure, respiratory distress, and even death. It infects humans through exposure to the urine of animals carrying these bacteria or environmental samples contaminated with urine of the infected animal. While leptospirosis has a worldwide distribution, it particularly affects countries with tropical and subtropical climates (Taniguchi and Póvoa 2019). It spreads easily during climate catastrophes, and extreme weather is a high-risk factor, as in the case of the leptospirosis outbreak in Puerto Rico following Hurricane Maria in 2017.
Tracking down the culprit
Facing the current global climate change, a research group in Okinawa, Japan (Sato et al. 2019) sought to develop new tools to systematically detect Leptospira in order to prevent human infection. This group studied the bacterial ecosystem that allows the development of Leptospira during biofilm formation and investigated which animals are potential reservoirs for transmitting these pathogens to humans.
|
|
  |
|
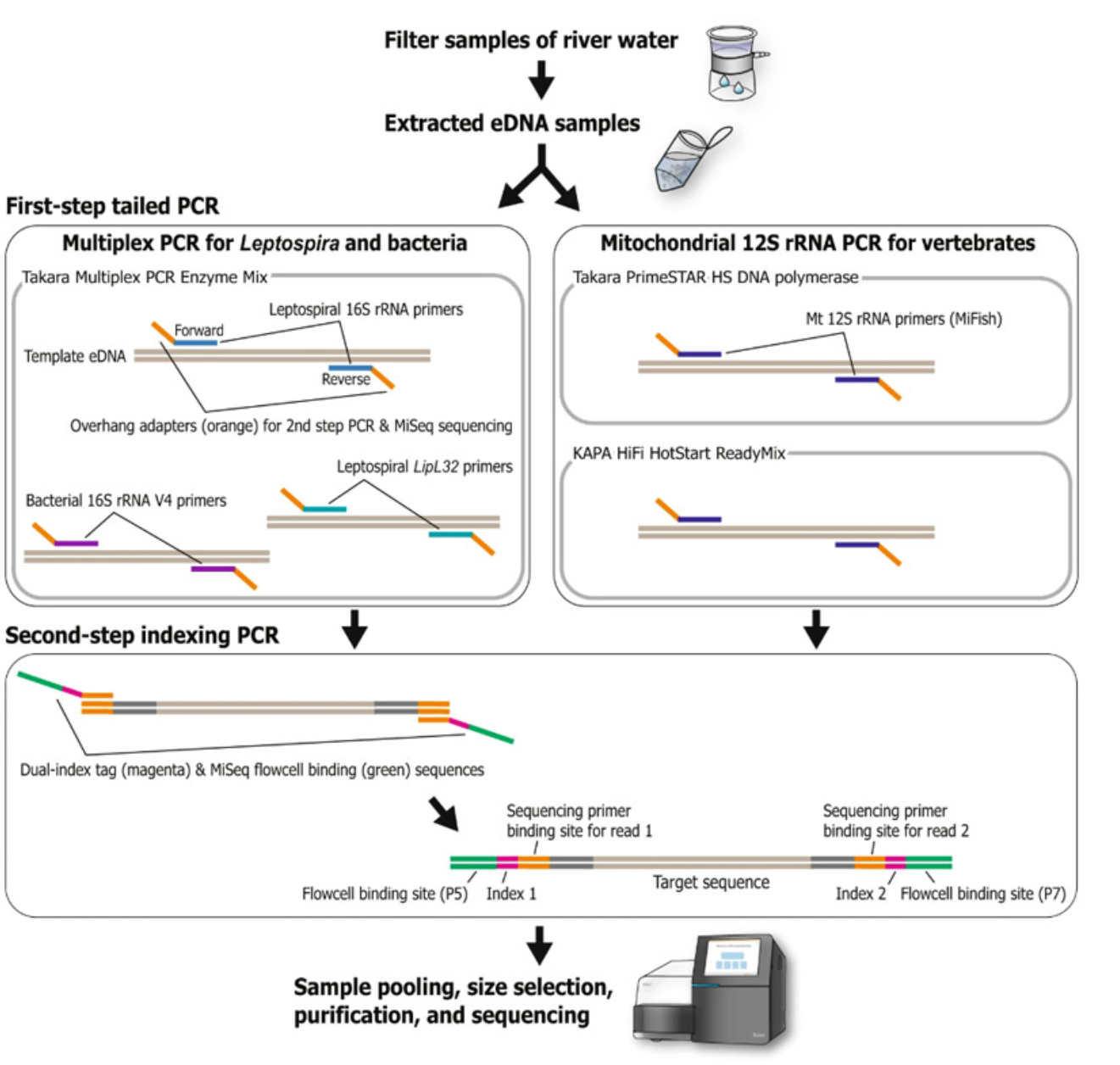
Figure 1. Schematic view of the library preparation procedure for metabarcoding sequencing based on a two-step tailed PCR. Multiplex PCR was applied in the first step for Leptospira and bacterial detection. The procedure for vertebrate mitochondrial 12S rRNA sequencing was basically the same but slightly modified from that of Miya et al. 2015. Image and caption adapted from Sato et al. 2019 and used under a Creative Commons Attribution 4.0 International License.
Why our polymerases are the investigative tools of choice
This study highlights the use of Takara Ex Taq HS and PrimeSTAR HS DNA polymerases to detect bacteria in environmental water samples, which are typically difficult to work with due to the presence of PCR-inhibitory contaminants. It demonstrated the robustness of these two enzymes for performing DNA amplification with samples which pose such challenges. Since screening assays can help prevent health disasters provoked by weather crises, which are occurring with greater frequency and severity due to climate change, having reliable reagents to power those assays will be crucial for getting ahead of future outbreaks.
References
High Prevalence of Deadly Bacterial Disease Found in Puerto Rico. Yale School of Medicine. https://medicine.yale.edu/news-article/20887/ |
If you enjoyed reading our articles, why not sign up to our blog mailing list? You'll get new articles straight to your inbox as they're released!